STEMvision™
For human cultures, three Analysis Packages have been designed for scoring and counting hematopoietic colonies produced by erythroid, myeloid and multi-potential progenitor cells in 14-day assays of CB, BM and MPB cultured in MethoCult™ Optimum media. A fourth Analysis Package is also available for counting the total number of colonies in 7-day assays of CB cultured in MethoCult™ Express medium.
For mouse BM cultures, three Analysis Packages have been designed to count total numbers of hematopoietic colonies produced by all combined myeloid and erythroid progenitors in M3434, myeloid progenitors only in M3534 and erythroid progenitors only in M3436.
Selection of at least one Analysis Package is required for each instrument purchase. Additional Analysis Packages and software add-ons to support 21 CFR Part 11 compliance are sold separately. Select your preferred Analysis Package and select Request Pricing for further information. Several purchase and leasing options are available.
For more information about Instrument Services including additional service packages and installation software please see our instrumentation overview.
Browse our Frequently Asked Questions (FAQs) on performing the CFU assay and explore its utility as part of the cell therapy workflow.
| Document Type | 产品名称 | Catalog # | Lot # | 语言 |
|---|---|---|---|---|
| Manual | STEMvision™ | 22000 | All | English |
Data
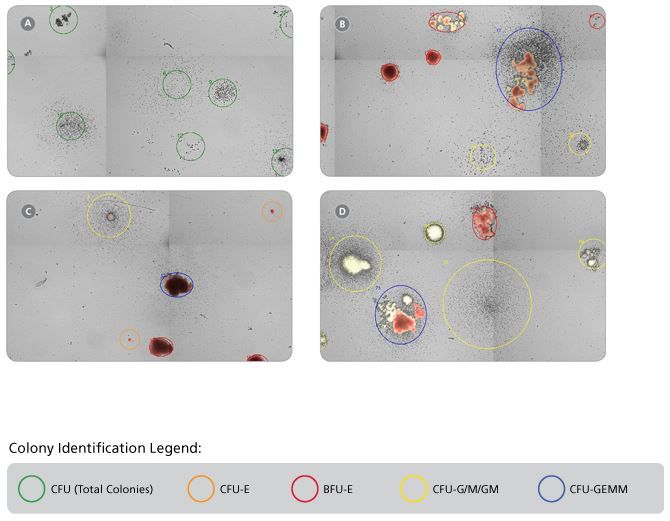
Figure 1. Representative STEMvision™ Images Showing Colonies Derived from CB Progenitors after 7 Days of Culture in MethoCult™ Express, and from CB, BM and MPB after 14 Days of Culture in MethoCult™ Optimum
These images have been analyzed by STEMvision™ Human (A) 7-Day and (B-D) 14-Day Analysis Packages. Green circles identify individual colonies in a 7-day CB CFU assay that scores total CFUs only (A). Orange and red circles identify erythroid colonies, yellow circles identify myeloid colonies and blue circles identify mixed colonies in 14-day CB (B), BM (C) and MPB (D) CFU assays. Erythroid and mixed colonies that contain hemoglobinized cells are shown in true red color.
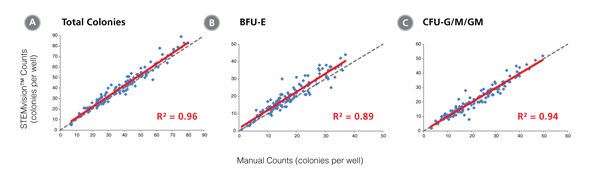
Figure 2. STEMvision™ Automated Counting of Total, Erythroid (BFU-E) and Myeloid (CFU-G/M/GM) Colonies Is Highly Correlated to Manual Counts of 14-Day CB CFU Assays
Cryopreserved CB cells were thawed, plated in MethoCult™ Optimum and cultured for 14 days. The resulting colonies were then counted both manually using an inverted microscope, and automatically using STEMvision™ with the Human CB 14-Day CFU Analysis Package (Catalog #22005). Gray dashed lines represent a perfect linear correlation between manual and automated counts. Red solid lines represent the actual linear correlation between manual and automated counts. The mathematical equations and correlation coefficients (R2) that describe each data set (n=130 CFU assays) are as follows: A: y=1.02x + 1.39, R2=0.96 for Total Colonies, B: y=1.05x + 1.53, R2=0.89 for BFU-E, C: y=0.99x + 0.13, R2=0.94 for CFU-G/M/GM.
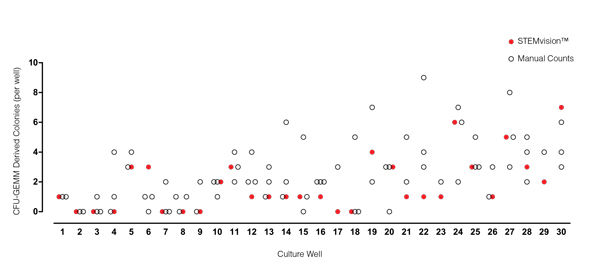
Figure 3. STEMvision™ Automated Counting of Mixed Colonies Falls Within the Range of Manual Counts of 14-Day CB CFU Assays
Thirty individual 14-day CB CFU assays were counted by three to seven people. The numbers of mixed colonies (CFU-GEMM) colonies counted manually in each well are shown by the black open circles (n=80 total assay scores). Manual CFU-GEMM counts in most cultures varied significantly between individual people. STEMvision™ counts of the same cultures (red circles) provided a CFU-GEMM count that was typically within the range of manual counts.
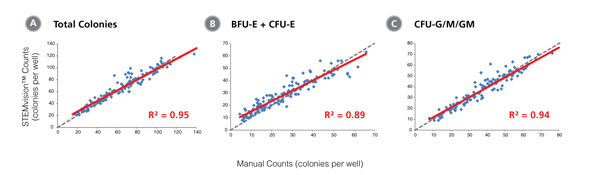
Figure 4. STEMvision™ Automated Scoring of Total, Erythroid (BFU-E + CFU-E) and Myeloid (CFU-G/M/GM) Colonies Is Highly Correlated to Manual Counts of 14-Day BM CFU Assays
Cryopreserved BM cells were thawed, plated in MethoCult™ Optimum, cultured for 14 days, and the resulting colonies then scored both manually using an inverted microscope and automatically using STEMvision™ with the Human BM 14-Day CFU Analysis Package (Catalog #22006). The BM Analysis Package can identify and count erythroid colonies produced by CFU-E and BFU-E separately, but these are combined in panel B. Gray dashed lines represent a perfect linear correlation between manual and automated counts. Red solid lines represent the actual linear correlation between manual and automated counts. The mathematical equations and correlation coefficients (R2) that describe each data set (n=120 CFU assays) are as follows: A: y=0.88x + 8.79, R2=0.95 for Total Colonies, B: y=0.83x + 6.71, R2=0.89 for CFU-E + BFU-E, C: y=0.92x + 2.55, R2=0.94 for CFU-G/M/GM.
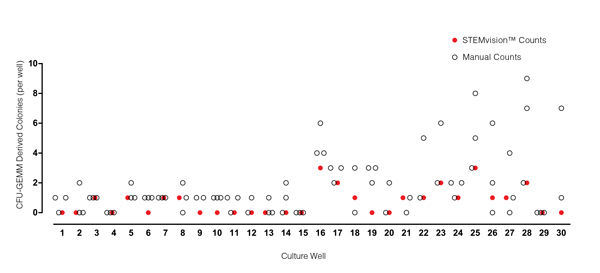
Figure 5. STEMvision™ Automated Counting of Mixed Colonies Falls Within the Range of Manual Counts of 14-Day BM CFU Assays
Thirty individual 14-day BM CFU assays were counted by three to seven people. The numbers of mixed (CFU-GEMM) colonies counted manually in each well is shown by the black open circles (n=82 total assay scores). Manual CFU-GEMM counts in most cultures varied significantly between individual people. STEMvision™ counts of the same cultures (red circles) provided a CFU-GEMM count that was typically within the range of manual counts.
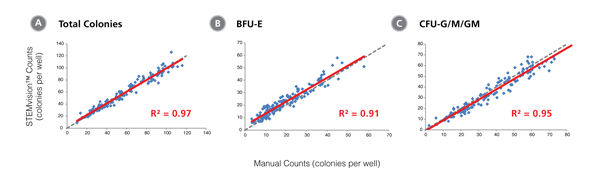
Figure 6. STEMvision™ Automated Counting of Total, Erythroid (BFU-E) and Myeloid (CFU-G/M/GM) Colonies Is Highly Correlated to Manual Counts of 14-Day MPB CFU Assays
Cryopreserved MPB cells were thawed, plated in MethoCult™ Optimum, cultured for 14 days, and the resulting colonies then scored both manually using an inverted microscope and automatically using STEMvision™ with the Human MPB 14-Day CFU Analysis Package (Catalog #22007). Gray dashed lines represent a perfect linear correlation between manual and automated counts. Red solid lines represent the actual linear correlation between manual and automated counts. The mathematical equations and correlation coefficients (R2) that describe each data set (n=143 CFU assays) are as follows: A: y=0.97x + 2.44, R2=0.97 for Total Colonies, B: y=0.96x + 3.74, R2=0.91 for BFU-E, C: y=0.96x + 0.90, R2=0.95 for CFU-G/M/GM.
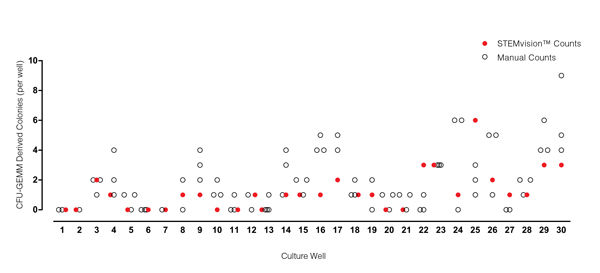
Figure 7. STEMvision™ Automated Scoring of Mixed Colonies Falls Within the Range of Manual Counts of 14-Day MPB CFU Assays
Thirty individual 14-day MPB CFU assays were counted by three to seven people. The numbers of mixed (CFU-GEMM) colonies counted manually in each well is shown by the black open circles (n=82 total assay scores). Manual CFU-GEMM counts in most cultures varied significantly between individual people. STEMvision™ counts of the same cultures (red circles) provided a CFU-GEMM count that was typically within the range of manual counts.
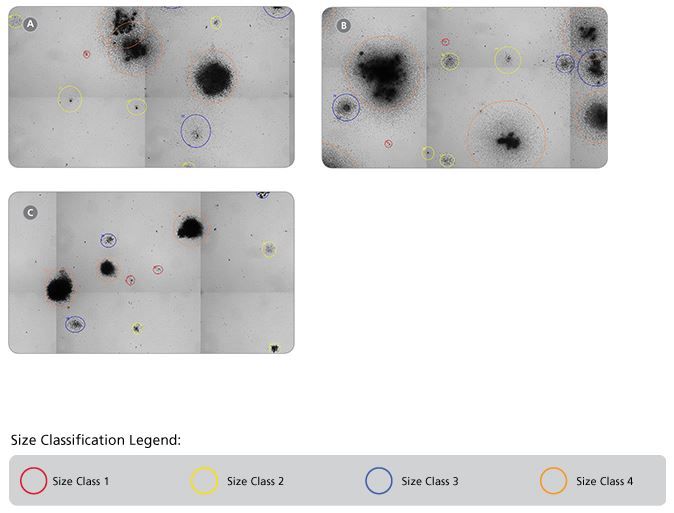
Figure 8. Representative STEMvision™ Images Showing Colonies Derived from Mouse BM Progenitors After 12 Days of Culture in MethoCult™ GF M3434, MethoCult™ GF M3534 or MethoCult™ SF M3436 Media
Images of mouse BM cells cultured in (A) MethoCult™ GF M3434, (B) MethoCult™ GF M3534 and (C) MethoCult™ SF M3436 were acquired using STEMvision™. The corresponding STEMvision™ Mouse Analysis Package (Table 1) was used to analyze each image. Red circles identify the smallest colonies - size class 1, yellow circles - size class 2, blue circles - size class 3 and orange circles identify the largest colonies - size class 4.
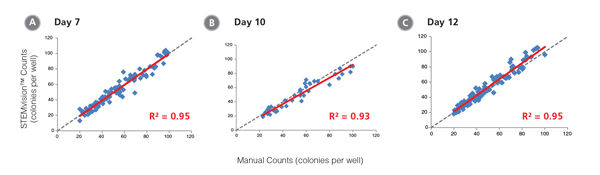
Figure 9. STEMvision™ Automated Counting is Highly Correlated to Manual Counting of Total (Myeloid Plus Erythroid) Colonies in Mouse BM CFU Assays
BM cells were plated in MethoCult™ GF M3434 (Catalog #03434/03444). Colonies were counted on days (A) 7, (B) 10 and (C) 12 both manually using an inverted microscope, and automatically using STEMvision™ equipped with the Mouse Total CFU Analysis Package (Catalog #22008). We recommend counting CFU assays of mouse progenitor cells plated in M3434 between 10 and 12 days. Gray dashed lines represent a theoretical perfect linear correlation between manual and automated counts. Red solid lines represent the actual linear correlation between manual and automated counts. The slope, 95% confidence intervals, correlation coefficients (R2) and sample size for each data set are provided in Table 1.
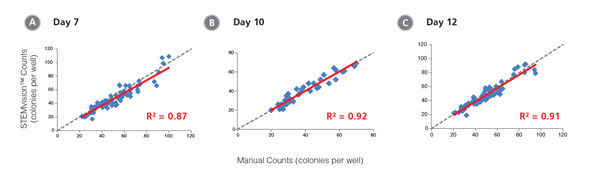
Figure 10. STEMvision™ Automated Counting is Highly Correlated to Manual Counting of Myeloid Colonies in Mouse BM CFU Assays
BM cells were plated in MethoCult™ GF M3534 (Catalog #03534). Colonies were counted on days (A) 7, (B) 10 and (C) 12 both manually using an inverted microscope, and automatically using STEMvision™ equipped with the Mouse Myeloid CFU Analysis Package (Catalog #22009). We recommend counting CFU assays of mouse myeloid progenitor cells plated in M3534 between 10 and 12 days. Gray dashed lines represent a theoretical perfect linear correlation between manual and automated counts. Red solid lines represent the actual linear correlation between manual and automated counts. The slope, 95% confidence intervals, correlation coefficients (R2) and sample size for each data set are provided in Table 1.
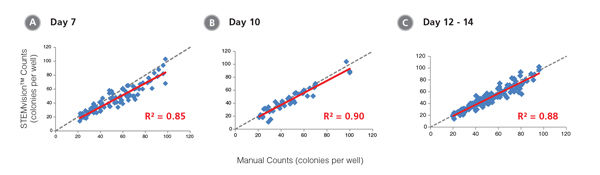
Figure 11. STEMvision™ Automated Counting is Highly Correlated to Manual Counting of Erythroid Colonies in Mouse BM CFU Assays
BM cells were plated in MethoCult™ SF M3436 (Catalog #03436). Colonies were counted on days (A) 7, (B) 10 and (C) 12 - 14 both manually using an inverted microscope, and automatically using STEMvision™ equipped with the Mouse Erythroid CFU Analysis Package (Catalog #22011). We recommend counting CFU assays of mouse erythroid progenitor cells plated in M3436 between of 10 to 14 days. Gray dashed lines represent a theoretical perfect linear correlation between manual and automated counts. Red solid lines represent the actual linear correlation between manual and automated counts. The slope, 95% confidence intervals, correlation coefficients (R2) and sample size for each data set are provided in Table 1.
Table 1. Correlation Between Automated STEMvision™ and Manual Colony Counting
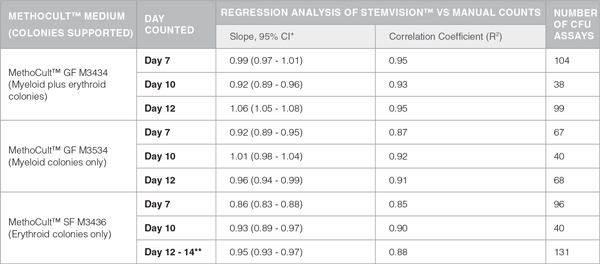
*CI: Confidence Internal
**Mouse CFU assays of erythroid progenitor cells plated in M3436 should be counted between 10 to 14 days.